Enzymes are biological catalysts, fundamental to the chemistry of life. Broadly, they facilitate conversion of substrates (reactants) into products at greatly accelerated rates. In this article, I develop general kinetic equations to describe a common means of enzymatic regulation: substrate inhibition. For comparison, it is useful to start by reviewing the traditional, most simplified kinetic model of enzyme kinetics. An enzyme must first bind the substrate (a process described by the Michaelis constant, Km). Next, it catalyzes the conversion of the substrate into the product (the rate constant for this process is the turnover number, kcat):
This kinetic scheme gives rise to the Michaelis-Menten equation:
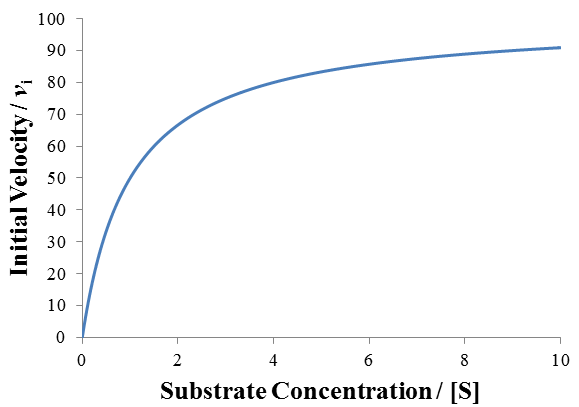
Where vi is the initial reaction rate, [E]T is the total enzyme concentration, and [S] is the substrate concentration. Often, the product kcat*[E]T is grouped together and termed the maximum velocity or “Vmax.” The parameter gives the maximum rate of substrate turnover, when the enzyme is saturated with substrate. Normally, increasing the concentration of the substrate increases the activity of the enzyme up to this maximum rate. This is because increasing the substrate concentration increases the proportion of enzymes which have substrate bound in their active site. The concentration of substrate required to saturate the active site of all enzymes in solution with the substrate depends on the binding affinity of the enzyme for the substrate. This can be seen clearly by plotting the Michaelis-Menten equation, as I have done below given Vmax = 100 and Km = 1.
In principle, maximum reaction rate and complete saturation of the enzyme with substrate occur only at infinite substrate concentration. However, once the substrate concentration is well above the Km value (for instance, when [S] > 10*Km) further increases in substrate concentration afford little increase in reaction rate, as most enzymes already have substrate bound.
Although many enzymes are described well by the Michaelis-Menten equation, many require more complex models. One such deviation is substrate inhibition. In this case, the enzyme must have at least two binding sites for the substrate. Typically, binding of the first substrate results in a catalytically active enzyme. However, if the enzyme is subject to substrate inhibition, the binding of an additional substrate to the other binding site results in a catalytically inactive enzyme. Where the goal of enzymes are to speed up the rates of reactions in biological systems, it may seem at first strange and counter-productive for an enzyme to be inhibited by its own substrate. This certainly has the potential to greatly reduce or completely halt catalysis. Biological systems are highly ordered, it is not sufficient for all reactions to always proceed as quickly as possible. Instead, the rates of reactions must be highly regulated to facilitate processes like homeostasis. Substrate inhibition is therefore used as an important regulatory mechanism used by a variety of enzymes to achieve different ends. For instance, phosphofructokinase is an important enzyme in glycolysis, a bioenergetically essential process where energy is liberated from glucose in the form of ATP. Although there is net gain of ATP over glycolysis, an essential step is the phosphorylation of fructose 6-phosphate using ATP. Phosphofructokinase catalyzes this conversion. Interestingly, phosphofructokinase shows substrate inhibition with respect to ATP. The regulatory function of this is clear: glycolysis is not needed if there is already sufficient abundance of ATP in the cell. When there is ample supply of ATP, substrate inhibition on phosphofructokinase blocks further glycolysis. Conversely, as ATP concentrations fall there is less substrate inhibition and the rate of glycolysis increases. This is merely one example, there are many well understood and distinct examples where substrate inhibition acts as a physiologically significant regulatory mechanism. For a more detailed discussion and additional examples see this article.
Although many enzymes are described well by the Michaelis-Menten equation, many require more complex models. One such deviation is substrate inhibition. In this case, the enzyme must have at least two binding sites for the substrate. Typically, binding of the first substrate results in a catalytically active enzyme. However, if the enzyme is subject to substrate inhibition, the binding of an additional substrate to the other binding site results in a catalytically inactive enzyme. Where the goal of enzymes are to speed up the rates of reactions in biological systems, it may seem at first strange and counter-productive for an enzyme to be inhibited by its own substrate. This certainly has the potential to greatly reduce or completely halt catalysis. Biological systems are highly ordered, it is not sufficient for all reactions to always proceed as quickly as possible. Instead, the rates of reactions must be highly regulated to facilitate processes like homeostasis. Substrate inhibition is therefore used as an important regulatory mechanism used by a variety of enzymes to achieve different ends. For instance, phosphofructokinase is an important enzyme in glycolysis, a bioenergetically essential process where energy is liberated from glucose in the form of ATP. Although there is net gain of ATP over glycolysis, an essential step is the phosphorylation of fructose 6-phosphate using ATP. Phosphofructokinase catalyzes this conversion. Interestingly, phosphofructokinase shows substrate inhibition with respect to ATP. The regulatory function of this is clear: glycolysis is not needed if there is already sufficient abundance of ATP in the cell. When there is ample supply of ATP, substrate inhibition on phosphofructokinase blocks further glycolysis. Conversely, as ATP concentrations fall there is less substrate inhibition and the rate of glycolysis increases. This is merely one example, there are many well understood and distinct examples where substrate inhibition acts as a physiologically significant regulatory mechanism. For a more detailed discussion and additional examples see this article.
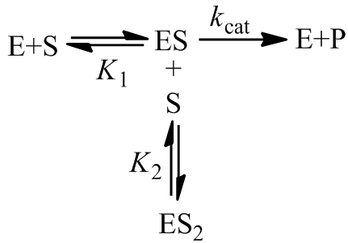
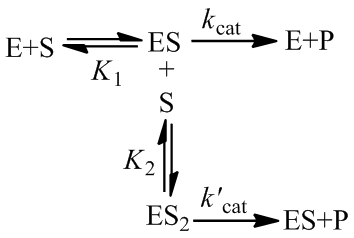
Classical Substrate Inhibition
In the “classical” kinetic model for substrate inhibition the enzyme has catalytic activity when one substrate is bound, but absolutely no activity if two are bound:
The kinetic equation corresponding to this kinetic scheme is:
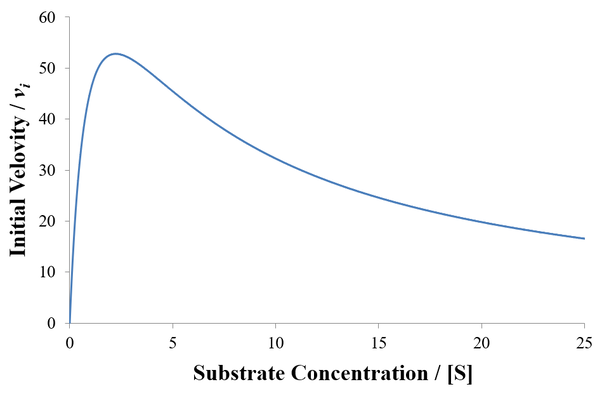
It is analogous to the Michaelis-Menten equation, but contains an extra term in the denominator to account for the catalytically inactive ES2 state. This has a dramatic effect on shape of the velocity curve, as at high substrate concentrations activity decreases due to occupancy of this state. An example is plotted below given Vmax = 100, K1 = 1, and K2 = 5.
A significant parameter in descripting substrate inhibition is the optimum substrate concentration, the concentration at which the enzyme has maximum activity. To calculate this we can utilize the fact that at maxima the derivative is zero:
Recall that a fraction equals zero when its numerator equals zero (given its denominator is nonzero). The optimum substrate concentration, [S]*, is therefore given by:
In this model the expression for the optimum substrate concentration is remarkably simple. It is the geometric mean of the two Michaelis constants corresponding to the first and second substrate binding event.
A Generalized Description of Substrate Inhibition
Despite the elegance of the above model, more complex substrate inhibition kinetic schemes are possible. In particular, the binding of the second substrate needn’t result in complete loss of activity. For many enzymes the second substrate molecule binding results in lower, but nonzero activity. I therefore develop the kinetic equation for the general case:
Note that this scheme allows the enzyme to have both higher or lower activity in when one or two substrate molecules are bound. Of course in the case that k'cat > kcat, this is no longer substrate inhibtion since the second binding event is actually activating. The enzyme may even be completely inactive in either the one or two substrate-bound state (either kcat or k'cat could be zero). Lastly, it is possible for the enzyme's affinity for binding the second substrate molecule to be either higher or lower upon binding the first. I derive the initial velocity equation for the kinetic scheme below:
General Equation for Optimum Substrate Concentration
I am unaware of any attempt in the literature to calculate optimum substrate concentrations for this more complicated, but more realistic kinetic model. Interestingly, in this model not all sets of parameters even result in there being an optimum concentration. I herein reveal what conditions are required for an optimum concentration to exist and present a novel equation to calculate it. As for the simpler case considered above, an optimum substrate concentration exists when there is a maximum velocity at a particular substrate concentration. Being a maximum, the derivative at this concentration must be zero.
To find the optimum substrate concentration, if one exists, we seek solutions to:
This condition is met when the numerator is equal to zero:
Notice that this is a quadratic equation. In general, the roots of a quadratic are given by:
Therefore:
The only valid solutions to this equation occur where the discriminant is subtracted, as shown. The reason for this is explained in detail below, along with the conditions required for there to be an optimum substrate concentration.
Intuitively, it is clear that [S]* is undefined when V2 > V1. In this case, binding of the second substrate molecule is activating and thus maximum activity is approached asymptotically towards infinite substrate concentration. Hence, there is no defined maximum. It is also possible to show this mathematically. Let us think back to the more general form of the solutions to the quadratic equation:
Intuitively, it is clear that [S]* is undefined when V2 > V1. In this case, binding of the second substrate molecule is activating and thus maximum activity is approached asymptotically towards infinite substrate concentration. Hence, there is no defined maximum. It is also possible to show this mathematically. Let us think back to the more general form of the solutions to the quadratic equation:
Since [S]* is a concentration it must be >= 0. Thus, negative solutions to this equation are not valid. Note that the b and c terms are always positive since the kinetics constants are always positive:
The sign of the a term depends on the difference between V1 and V2. If V2 > V1, the a term is positive. If V2 < V1, then it is negative. When the a term is positive, both solutions give negative values. However, when the a term is negative, subtracting the discriminant must give a positive solution.
In conclusion, V1 > V2 is a necessary condition for there to be an optimum substrate concentration. Furthermore, notice also that when this condition is met the discriminant must be positive and so for any V1 > V2 there is a solution, independent of the values of the other constants. In this case the term must negative and so the discriminant will always be.
Special Cases of the Substrate Inhibition Equation
Special case I: ES is catalytically active, ES2 is inactive
In the special case where the enzyme bound to a single substrate is active, but completely inactive when two substrate molecules are bound (that is, V1max > 0, V2max = 0). Of course the general equation simplifies to give the markedly simplified result obtained earlier.
In the special case where the enzyme bound to a single substrate is active, but completely inactive when two substrate molecules are bound (that is, V1max > 0, V2max = 0). Of course the general equation simplifies to give the markedly simplified result obtained earlier.
Special case II: ES is catalytically inactive, ES2 is active
In the special case where the enzyme bound to a single substrate is inactive, but active when two substrate molecules are bound (that is V1max = 0, V2max > 0). Formation of the ES complex has the effect of decreasing the “apparent Km” of the enzyme, it partitions a fraction of the enzyme population into an inactive state in the presence of substrate. This effect is not truly simply increasing the Km value though, as the situation is no longer classical Michaelis-Menten kinetics:
In the special case where the enzyme bound to a single substrate is inactive, but active when two substrate molecules are bound (that is V1max = 0, V2max > 0). Formation of the ES complex has the effect of decreasing the “apparent Km” of the enzyme, it partitions a fraction of the enzyme population into an inactive state in the presence of substrate. This effect is not truly simply increasing the Km value though, as the situation is no longer classical Michaelis-Menten kinetics:
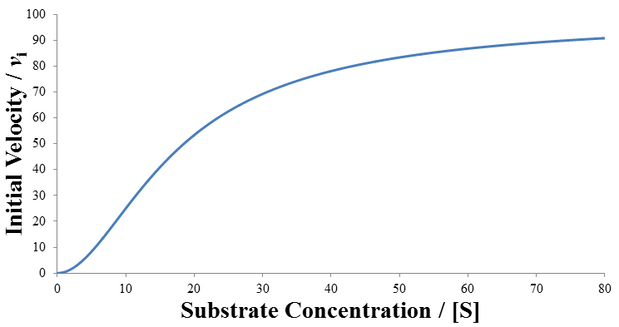
Comparing this to the Michaelis-Menten equation, we see that there are two substrate-dependent terms in the denominator. The alternative behavior of this system compared to normal Michaelis-Menten kinetics is most clear visually. Observe below where the initial velocity is plotted as a function of substrate concentration given V1 = 0, V2 = 100, K1m = 50, and K2m = 5. Note that the curve’s shape is now sigmoidal instead of a rectangular hyperbola.
Nevertheless, it is insightful to describe these kinetics in terms of something analogous to Km. This allows us to study the effect that the inactive ES complex has on the kinetics of the enzyme. Under Michaelis-Menten kinetics, Km represents the substrate concentration required to achieve 50% of the maximum enzyme activity. Thus, by analogy we can calculate the concentration required to reach half of maximum activity ([S]1/2):
Interestingly, lowering the affinity of the enzyme for the first binding event (increasing K1m) actually increases [S]1/2, meaning that it increases the concentration of substrate required to saturate the enzyme into its active state with two substrates. This is because binding of substrates is sequential and the enzymes must bind the first substrate before binding the second, even if the enzyme’s affinity for the second substrate is greater than the first.
Special Case III: No Cooperativity (K1 = K2 = Km)
In this case, the enzyme exhibits no cooperativity with respect to substrate binding; binding the first substrate has no effect on the enzyme’s affinity for the second. Furthermore, the binding sites for the two substrates would likely be symmetric. By making the substitution K1 = K2 = Km, we obtain a simplified equation for the optimum substrate concentration:
Special Case III: No Cooperativity (K1 = K2 = Km)
In this case, the enzyme exhibits no cooperativity with respect to substrate binding; binding the first substrate has no effect on the enzyme’s affinity for the second. Furthermore, the binding sites for the two substrates would likely be symmetric. By making the substitution K1 = K2 = Km, we obtain a simplified equation for the optimum substrate concentration:
What is most striking about this result is that the optimum substrate concentration is now directly proportional to the Michalis constant, Km. This simplification results from the unique symmetry of this special case. As Km is increased the enzyme’s propensity to be in the one and two substrate-bound state decrease symmetrically so that there is a linear increase in [S]*.
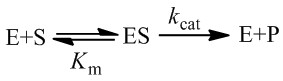
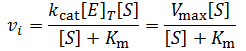
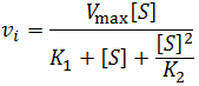
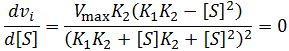
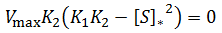
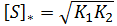
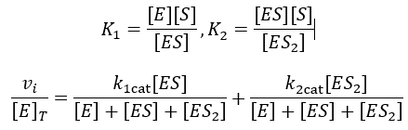
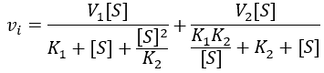
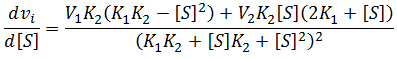
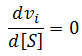
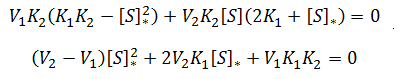
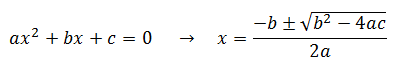
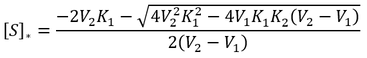
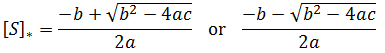
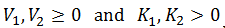
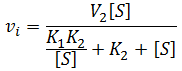
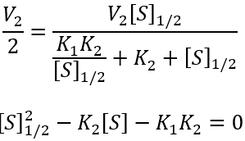
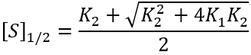
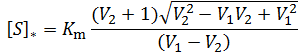
 RSS Feed
RSS Feed